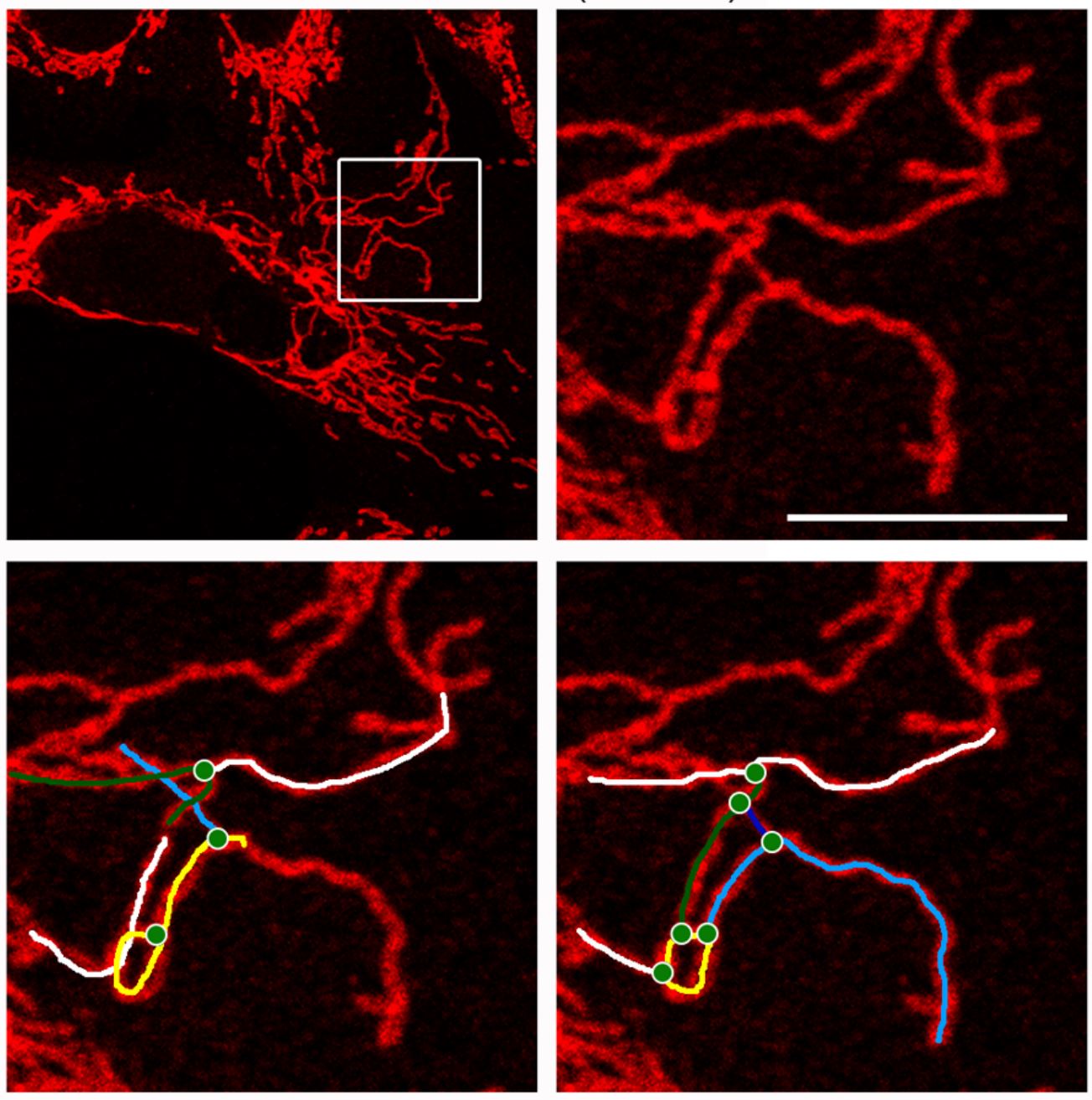
Decoding mitochondrial networks through confocal imaging.
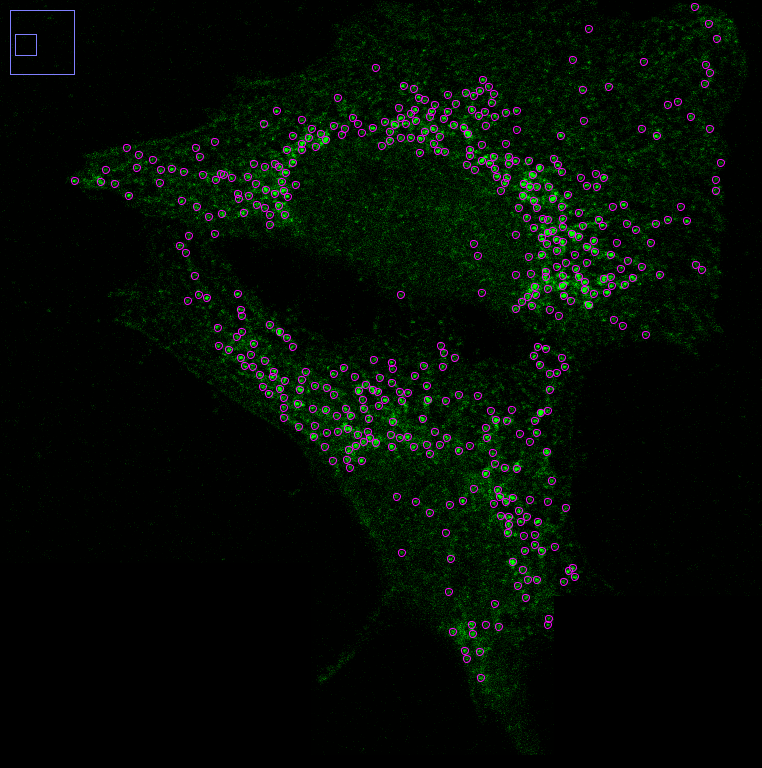
Mitochondrial DNA (mtDNA) nucleoids.
Mitochondria are highly complex organelles that play numerous roles in the cell, far beyond their popular description as the "powerhouse of the cell." These dynamic structures form interconnected networks whose size and topology adapt to the metabolic needs and physiological state of the cell. To better understand these networks, we developed methods to analyze their architecture using confocal microscopy, allowing us to visualize and describe their intricate structures. These networks are not static; they constantly undergo fusion and fission events, responding to cellular conditions and contributing to various processes such as energy production, signaling, and apoptosis.
Originating from an ancestral symbiotic relationship with a prokaryotic organism, mitochondria retain their own genome, known as mitochondrial DNA (mtDNA). This unique evolutionary heritage makes the regulation of mtDNA within the mitochondrial network a critical aspect of cellular function. To investigate this, we analyzed the positioning of mtDNA within the network and studied how network dynamics influence mtDNA maintenance and activity, as well as how mtDNA contributes to the network's structural and functional behavior. Understanding these interdependencies sheds light on the broader roles mitochondria play in cellular homeostasis and disease.

Decoding mitochondrial networks through confocal imaging.

Mitochondrial DNA (mtDNA) nucleoids.
[1] Ouellet, M., Guillebaud, G., Gervais, V., Lupien St-Pierre, D., & Germain, M. (2017). A novel algorithm identifies stress-induced alterations in mitochondrial connectivity and inner membrane structure from confocal images. PLOS Computational Biology, 13(6), e1005612.
[2] Patten, D. A., Ouellet, M., Allan, D. S., Germain, M., Baird, S. D., Harper, M.-E., & Richardson, R. B. (2019). Mitochondrial adaptation in human mesenchymal stem cells following ionizing radiation. The FASEB Journal, 33(8), 9263.
[3] Ilamathi, H. S., Ouellet, M., Sabouny, R., Desrochers-Goyette, J., Lines, M. A., Pfeffer, G., Shutt, T. E., & Germain, M. (2021). A new automated tool to quantify nucleoid distribution within mitochondrial networks. Scientific Reports, 11(1), 22755.
[4] Vishwakarma, A., Chikhi, L., Todkar, K., Ouellet, M., & Germain, M. (2024). Mitochondrial Dysfunction alters Early Endosome Distribution and Cargo Trafficking via ROS-Mediated Microtubule Reorganization. BioRxiv, 2024–08.
The Curse of Endless Links.